Searching for clues to the next pandemic
By Nicole Lim, Senior editor
A colony of bats that roosts in La Ang Pracheav cave in Battambang Province takes flight to forage at dusk (left); the scientists work late into the night to complete the sampling // Credit: Sophie Borthwick
Around mid-afternoon on a Friday, a convoy of mud-splattered four-wheel-drive pickup trucks pulled to a halt at the edge of a field surrounded by shrubbery, some 250km north-east of Cambodia’s capital Phnom Penh. The doors pushed open and a team of researchers uncrumpled into the open.
In this issue of MEDICUS—the Podcast, Sophie Borthwick, Dolyce Low and Lena Ch’ng talk about their work and what keeps them motivated (12-minute listen). Listen to more MEDICUS - the Podcast episodes on Apple Podcasts, Google Podcasts and Spotify.
It was 7 February 2020 and the team were on what would turn out to be their last trip to Cambodia for their genomic surveillance study.
“It was surreal—the end of an era,” said Sophie Borthwick looking back. “And sad that I wouldn’t get to see the team all the time and continue the work that I love to do but happy at the huge feat that we had accomplished and excited about the science we still had to do back in the labs,” she added.
Borthwick, a senior research associate in the Laboratory of Virus Evolution at Duke-NUS, had been shuttling to and from Cambodia almost every other month for three years as she and the team travelled up and down the country to collect samples from bats and small mammals—two key virus reservoirs—to find out what threats could be hiding in these creatures.
With the fieldwork almost completed, the team is now focusing on supporting their Cambodian counterparts in finishing the analysis of the almost 30,000 samples that they had gathered from 1,048 small mammals and 2,460 bats.
“We had planned to go back and help the staff finish the lab work and organise a big party with the field and lab teams to celebrate a successful project,” mused Borthwick, a senior research associate from Duke-NUS.
But all plans had to be shelved due to just the kind of threat her work hopes to nip in the bud: the COVID-19 pandemic.
Despite the last-minute spanner, Borthwick and her colleagues, together with their Cambodian and other international partners, have broken new ground in genetic surveillance. Having travelled to 227 out of the 250 randomly identified sites across 24 of Cambodia’s 25 provinces, their study became by far the most extensive genomic surveillance effort undertaken to date.
“What the COVID-19 pandemic has shown us is how important active field surveillance and biological sample collection are,” said Dolyce Low, another member of the Laboratory for Virus Evolution. “It showed us how little we know about the viral and bacterial communities harboured by bats and other disease reservoirs like rodents.”
With a new statistical analysis from Duke University highlighting that pandemics are more likely than most people think, work like this will be key to give humanity a fighting chance or possibly even the upper hand.
When the holes in the Swiss cheese align
While incursions from zoonotic viruses are fairly common, most hit a dead end. Some are simply too deadly, killing their new host so fast that the host has no chance to pass the virus on to anyone else. Some are not well-adapted to the human body and struggle to cause an infection before the immune system kicks them out.
“Many spillover events don’t become epidemics because the virus doesn’t develop the necessary mutations,” said Low, adding that in their work, the team has found signs of past infection with a filovirus—a diverse family of viruses whose most infamous member is Ebola—in bat hunters from Northeast India.

A sampling station set up along the cliff edge outside the bat cave in October 2018 (left) // Credit: Dolyce Low
“We found the same antibodies in the bats as we did in the bat hunters, which shows that the virus jumped into humans, but it didn’t develop into something that could cause human disease or death,” said Low.
But every now and again, one virus will slip through. The most prevalent strain of the human immunodeficiency virus has been traced to what appears to be one single spillover event that took place along a minor tributary of the mighty Congo River around about 1908. And based on the statistical analysis from a team at Duke, in any given year, humans have a two per cent chance of having to square up against a potential COVID-like pandemic.
While the purpose of the Duke study was not to describe why these pandemics occur, co-author William Pan, an environmental health scientist, believes that the answers lie in how humans interact with each other and their environment.
“Studying the environment, climate change, our food systems and how they are connected is all part of understanding why diseases emerge and spread rapidly through populations,” said Pan, who is an associate professor of global environmental health at Duke. “And it is something that we need to get a handle on fairly quickly.”
To get a handle on this intricate dynamic, genomic surveillance of key virus reservoirs at high-risk interfaces—particularly in some of the most biodiverse regions of the world—is key.
“Those are tropical forests, where there’s rainfall, where we already know there’s a lot of diversity in terms of animal and plant life, where we know humans live on the edges of large forests—those types of locations need to strengthen genomic surveillance and public health in general,” said Pan.
Places like guano farms present ideal opportunities for a virus to jump from an infected animal to a human through a small cut or wound // Credit: Sophie Borthwick
Precisely the sort of places that Borthwick and Low, along with the rest of the team, have been travelling to as they survey what threats lie just beyond the human sphere.
Because bats are found on all inhabited continents and are one of the most diverse mammalian species—with some 1,400 species—they carry a similarly rich diversity of microscopic threats.
“By going to these different locations, we can determine where specific viruses and bacteria are hiding,” said Lena Ch’ng, a research assistant who recently re-joined the lab after interning with them first as a teenager.
By examining the exchange of genes between bats and small mammals—another top virus reservoir—that live in different habitats, the team can spot areas of potential spillover into humans.
“These spillovers are particularly likely at high-risk interfaces, which include bushmeat hunting, guano farming and live animal trading,” added Ch’ng.
More than just a field trip
The team collects a wide range of biological samples. The samples are stored in cryotubes—which are pre-sorted by the colour of their caps, with each colour representing a different sample type—and put on ice for the journey back to the local lab, where they will be analysed by the laboratory team.
Samples are stored in cryotubes and sorted by colour, with each colour identifying a different type of tissue, (left) and later sorted by colour into boxes and stored in ice-filled cooler boxes (right) // Credit: Sophie Borthwick
But they do more than work alongside each other. A key pillar of the work for the Duke-NUS team is to ensure that surveillance can carry on long after they have packed up and returned to Singapore. From donning PPE and fitting N95 masks to collecting samples safely and optimising protocols, the team makes sure that their counterparts are trained in all.
“We train our in-country counterparts so that they can carry on independently after our project is completed. That way, they can pursue the questions that are most pressing on their public health agenda and, in turn, train the generation after them,” said Borthwick.
The team have trained teams not just in Cambodia but also in India, Kazakhstan and Singapore. Back in February 2020, they were about to embark on a similar project in Laos just as the pandemic closed borders and halted travel.
A scientist in Kazakhstan prepares to collect nasal swabs from dromedary camels (left); a training session on how to safely sample bats (centre); and field sampling volunteers practise how to collect environmental samples from under bat roosts in August 2019, before all fieldwork was suspended from the pandemic (right) // Credit: Ian Mendenhall (left),Benjamin Lee (centre) and Dolyce Low (right)
In pursuit of science from field to lab
While the team relishes the hot and sweaty fieldwork, what makes them so passionate about their work is that they can see the project through from collection to analysis.
“It is really rewarding when you finally get to the end of your analysis and make a phylogenetic tree,” said Borthwick.
Low added, “Especially when you know exactly where you collected that sample that now sits on a tree in a peer-reviewed paper has come from!”
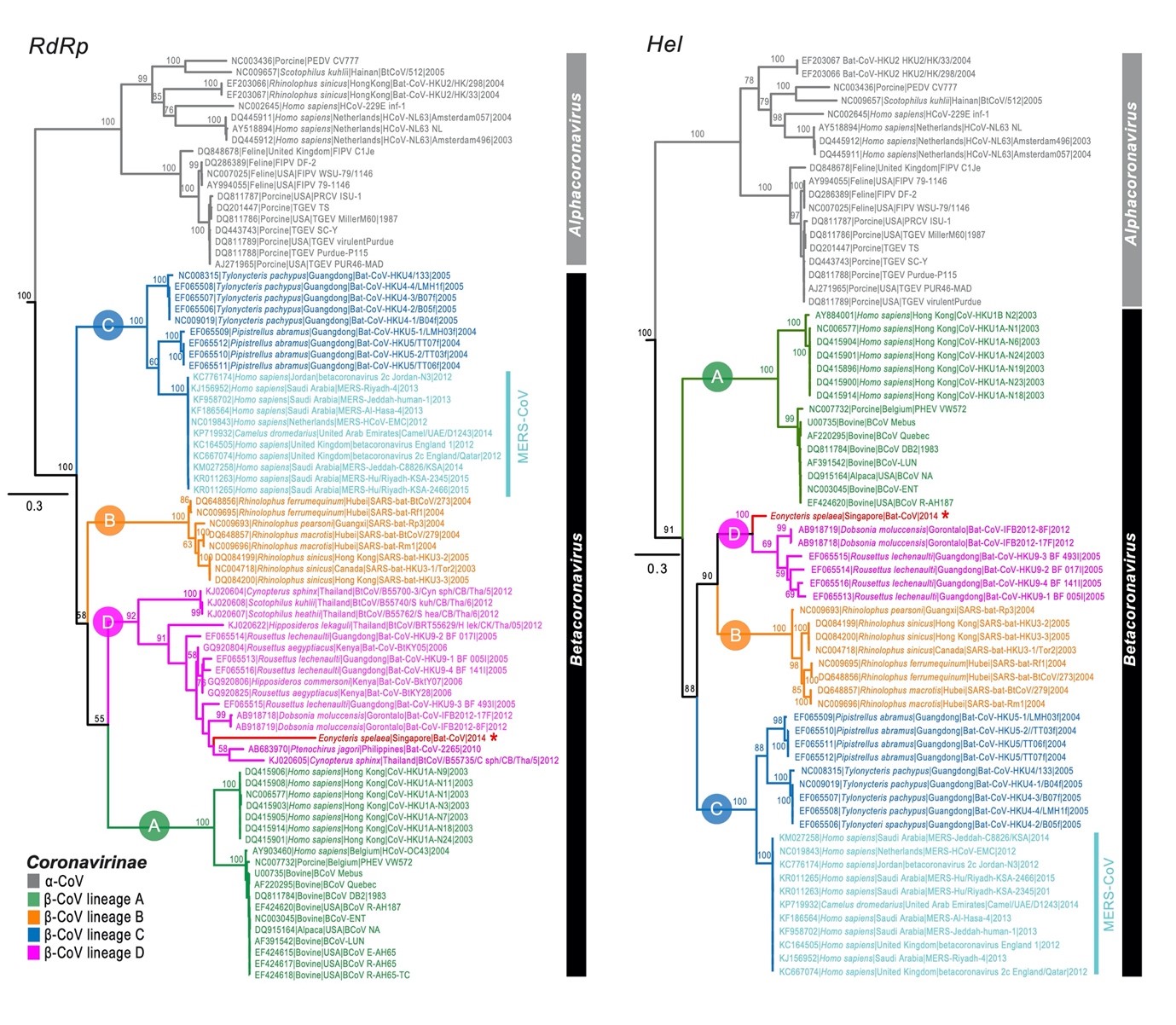
This phylogenetic tree of the RdRp and Hel nucleotide sequence of alpha- and beta-coronavirus published in 2016 includes newly idenfitied coronavirus sequences (marked with an asterisk) collected from cave nectar bats // Source:
doi/10.1111/tbed.12568
But deciphering what’s in a sample isn’t like diagnosing an infection in humans.
“There is a significant difference between clinical samples, which are taken in clean settings, and environmental samples, which are collected in the field. We don’t know when the animals are shedding and don’t have a good idea if the animals we catch are ill, unlike on the human side where it’s easier to establish that a human is ill,” said Ch’ng, who added that though the uncertainty of viral load—particularly in faecal samples—may be frustrating at times, the team gets a sense of what’s good to eat.
“Based on the stool samples’ changes in colour over time, we can spot that this is the season for papaya, that’s the season for berries,” said Ch’ng.
Because they are not conducting a diagnostic test, which detects a specific virus like Influenza A, the team screens each sample for several families of viruses and bacteria to make sure that they won’t miss any of the usual suspects.
“We might start by screening for as many as ten viruses,” explained Low.
They use high-throughput technologies like next-generation sequencing to rapidly sequence any scraps of genetic material, such as DNA or RNA, that are floating in the sample. The team instructs the software to focus on the parts of the virus that don’t mutate as quickly, which they tag so that they can rapidly search and filter their gigabytes of data.
With a hint of a virus family revealed, the team then has to tunnel down to find out what strain they have in their sample.
“At times, we have to try multiple methods before we get our answer,” said Ch’ng. “You almost start to doubt yourself during those moments.”
And during the pandemic, this has been particularly challenging as the Duke-NUS team had to support their Cambodian counterparts remotely. They didn’t just have to overcome language barriers but also battle malfunctioning equipment or incompatible software that made troubleshooting almost impossible.
“Doing the experiments with our own hands can be challenging as we have to tweak many things along the way, so putting ourselves in their shoes and trying to communicate what they need to do over Zoom was extra challenging,” said Ch’ng.
Better armed to face tomorrow
Just as the lab experiments end with a set of new data characterising what bacteria and viruses are present in different bat species living in different habitats, the fieldwork comes to an end—usually in the deep of the night. When all the samples have been collected, labelled and stored in iceboxes.
The stations are packed up and the tired team, exhausted, sweaty and with N95 lines imprinted on their faces, heads back to the SUVs and begin the long journey home.
One of the 227 field trips the Duke-NUS team, their international colleagues and Cambodian counterparts undertook as part of the largest genomic surveillance effort in bats and small mammals to date